Identification of Post Harvest Fungi Associated Telfairia occidentalis Hook F. Leaves Using Basic Molecular Techniques
| Received 22 Apr, 2024 |
Accepted 22 Nov, 2024 |
Published 23 Nov, 2024 |
Background and Objective: The leaf of Telfairia occidentalis, a highly nutritious and economically important vegetable in Nigeria, is threatened by fungal diseases that affect its quality and yield. This study was conducted to isolate and identify the fungal species associated with the leaves of Telfaira occidentalis using both conventional microbiological techniques and molecular methods. Materials and Methods: Fungal species were isolated from Telfairia occidentalis obtained from the Botanic Garden of the University of Port Harcourt, Choba Port Harcourt, Rivers State, Nigeria. Potato dextrose agar (PDA) was used as a growth medium for the fungi. Deoxyribonucleic Acid (DNA) was extracted from pure cultures of fungal isolates using Quick-DNA Fungal/Bacteria Mini Prep Kit. Polymerase Chain Reaction (PCR) amplification of the internal transcribed spacer (ITS) region of the fungal isolates was carried out using fungal universal primer pairs ITS4 and ITS5. Sanger sequencing was performed and the sequences obtained were aligned and compared with sequences on the National Centre of Biotechnology Information (NCBI) database. Results: The result of the nucleotide sequence analysis revealed that isolate 1 is Trichorderma reesei with 590 base pairs and 99.66% similarity while isolate 2 is Aspergillus aculeatus with 610 base pairs and 99.62% similarity. A phylogenetic tree was constructed using the best BLAST hits. The evolutionary tree was inferred using the neighbor-joining method. Conclusion: The study identifies Trichoderma reesei and the newly discovered Aspergillus aculeatus on post-harvest Telfairia occidentalis leaves, raising concerns about potential health risks from consuming contaminated leaves due to harmful metabolites.
| Copyright © 2024 Gloria et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Green leafy vegetables provide crucial minerals and trace elements that contribute to the proper effectiveness of the body system by sustaining regular metabolic processes in restoring damaged cells, necessary maintenance and prevention of diseases in humans1. The leafy vegetable, Telfairia occidentalis Hook F., commonly called fluted pumpkin, is native to Nigeria and is also found in the moist coastal areas of West Africa but rarely occurs naturally in East Africa2. It is a warm-weather crop that grows well in lowlands and tolerates elevation of a few meters above the ground. It thrives best in soils rich in organic matter3. Fluted pumpkin belongs to the family Cucurbitaceae and is a strong climber and short-term perennial4. The leaves are harvested by pruning with a knife and harvesting by hand picking damages the plants and hinders the development of side shoots5.
This vegetable crop T. occidentalis has gained significant importance in its dietary practices and is now a prevalent component in regional households6. It is cultivated for its leaves and seeds which are good sources of organic acid, vitamins, proteins, oils and carbohydrates7. Telfairia occidentalis seeds and leaves are consumed because they are healthy sources of lipids, vitamins, fiber and minerals such as iron, potassium, phosphorus and mineral salt8. The fluted pumpkin leaf also contains 1.35% alkaloid, 1.50% flavonoid, 3.60% saponin, 12.20% phenol and 0.437% tannin9. Fluted pumpkin leaf contains iron which is usually extracted and given as blood tonic to patients lacking blood while the seeds are reportedly used for the treatment of arthritis10,11. The vegetable contains photosynthetic pigments called chlorophylls and carotenoids12. The study of Iweala and Obidoa13 reveals that the long-term feeding of T. occidentalis supplemented diet caused a significant increase in the weight of animals which may be due to its content of rich nutrients.
The most important losses in agricultural production involving the greatest farm economy costs occur by postharvest diseases. For example, in developing countries like Nigeria, over 85% of food consumed is obtained from the farm and the inability of food to be available at all seasons is affected by postharvest losses, this unavailability of food at all seasons causes food insecurity. Despite the nutritive and market value of T. occidentalis, sustainable production is greatly constrained by various diseases each year, of which leaf spot causal agent Phoma sorghina14. Molecular-based technologies are the most reliable tools for characterizing microorganisms as they deal with the genetic composition of organisms. Molecular tools have made it possible to obtain in-depth information on analyses of systems subject to climate change15, foodstuffs, agriculture, industrial settings and across the environmental sciences16. This study was therefore aimed at isolating and identifying fungal species associated with fluted pumpkin leaves using conventional microbiological techniques and molecular methods.
MATERIALS AND METHODS
Study area and sample collection: This study was carried out at the Regional Centre for Biotechnology and Bioresources Research Laboratory, University of Port Harcourt from January to February 2023, where the isolation of the organisms, extraction of DNA and amplification of Genomic DNA were carried out. The leave samples were collected in April from the Botanic Garden of the University of Port Harcourt, Choba Port Harcourt, Rivers State, Nigeria. The 10 samples were collected using a sterilized trowel and were transferred to clean, dry and labeled plastic containers. The samples were authenticated at the Regional Centre for Biotechnology and Bioresources Research and used for further analysis.
Isolation of fungal organisms
Serial dilution: All glassware used was autoclaved at 121°C for 15-20 min. The work area was sterilized with 70% ethanol. Several dilution blanks were prepared, transferred to test tubes and labeled 10–1, 10–2, 10–3, 10–4 and 10–5. They were separately filled with 9 mL of water except the first test tube labeled 10–1 which served as the stock solution and was filled with 10 mL of water. One gram of the leaf sample was transferred into the first test tube (10–1) and mixed properly; this was to aid even distribution of the sample in the tube. A micropipette was used to transfer 1 mL of the sample into the next test tube (10–2) and mix. One milliliter of the dilution was transferred from a test tube (10–2) into (10–3) and 1 mL was aseptically transferred from test tube 10–3-10–4. Finally, 1mL of the dilution from the test tube containing 10–4 dilution was transferred to the test tube labeled (10–5). The test tube labeled with 10–3 was used for further investigation.
Inoculation and incubation of fungal organisms: After making a dilution of leaf sample, 0.5 mL of the desired dilution (10–3) was pipetted onto the surface of freshly prepared PDA in a Petri dish. A sterilized bent glass rod was used to spread the sample evenly. Each Petri dish was well labeled with dates and sealed with masking tape.
The plates were incubated at room temperature (27±2°C) for 7 days under a laminar flow (ESCO Technologies Inc., Horsham, Pennsylvania, USA.) to obtain the fungal organisms. Fungal organisms when obtained were sub cultured to obtain pure cultures of fungi. Pure cultures were stored at 4°C in a refrigerator till when needed.
Extraction of fungal DNA: The DNA was extracted using the protocol of Zymo Quick-DNA Fungal/Bacterial Mini-Prep Kit (Zymo Research Group, California and USA), the protocol of the kit mentioned above was used with modifications.
The fungal mycelium was scrapped off from the surface of the plates using a sterilized surgical blade and transferred into a sterilized mortar and pestle. Bashing bead buffer (750 μL) was added to the sample before homogenizing with liquid nitrogen (-196°C). Homogenized samples were transferred into Eppendorf tubes of 1.5 mL and promptly put on ice. The Eppendorf tubes containing the homogenized samples were centrifuged in a refrigerated centrifuge at 10,000 g for 1 min. About 40 μL of the supernatant was transferred to a Zymo-Spin III-F Filter in a collection tube and centrifuged at 7,000 g for 1 min. The Zymo-Spin III-F Filter was discarded after use. About 1,200 μL of Genomic Lysis Buffer was added to the filtrate in the collection tube and properly mixed, 800 μL of the mixture was then transferred to a Zymo-Spin IIC Column in a collection tube and centrifuged at 10,000 g for 1 min. The flow-through was discarded from the collection tube and step 6 was repeated. About 200 μL of DNA Pre-Wash Buffer was added to the Zymo-Spin IIC Column and centrifuged at 10,000 g for 1 min. About 500 μL/g DNA of Wash Buffer was added to the Zymo-Spin IIC Column and centrifuged at 10,000 g for 1 min. The Zymo-Spin IIC Column was transferred to a clean 1.5 mL microcentrifuge tube and 50 μL of DNA Elution Buffer was added directly to the column matrix and centrifuged at 10,000 g for 30 sec to elute the DNA.
Determination of DNA concentration and purity using NanoDrop: DNA Concentration and purity were measured using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, Delaware, USA). Purity is measured as a ratio of absorbance at 260 nm to that of 280 nm. The Nanodrop was connected to a computer system and the sensor was sterilized using 70% ethanol, 1 μL of Elution buffer was dropped directly on the NanoDrop sensor to create a blank. The DNA samples were loaded separately and wiped after obtaining the result to avoid contamination.
Determination of quality of the DNA using gel electrophoresis: The gel electrophoresis allows the movement of samples from the cathode (-) to the anode (+) charge through agarose gel connected to a power source. To check the quality of the DNA, gel electrophoresis was performed. Half a gram (0.5 g) of agarose powder was mixed with 50 mL of Tris Boris EDTA (TBE) IX in a measuring flask and microwaved for 2 min to get a clear solution. A 3 μL of EZ-vision gel (blue light) was added to the content in the conical flask and then poured into the casting tray. The comb was inserted for the creation of wells. The content was allowed to sit between 20-30 min at room temperature for the gel to solidify. The gel electrophoresis machine (C.B.S. Scientific EPS-300X (C.B.S. Scientific Company, California, USA) was set up and the plate holding the gel was put into the tank. The TBE IX was poured into the gel tank until the gel was completely submerged. Each DNA sample which was mixed with 3 μL of EZ-vision gel blue light 2X was loaded separately into the wells made by the comb. The setup was allowed to run for 40 min. At the end of the run, the DNA fragments were viewed with a UV transilluminator (Lab net International, Edison, New Jersey, USA). Where bands appeared, the DNA was said to be of good quality.
Fungal DNA amplification and sequencing: The primers used for the fungal PCR were: ITS4- forward (5'TCCTCCGCTTATTGATATGS-3') and ITS5: Reverse (5'GGAAGTAAAAGTCGTAACAAGG-3'). The PCR was done in a total volume of 25 μL containing 2.5 mL of 10X PCR buffer, 1 μL of 25 mM MgCl2, l μL each of 5 μM forward and reverse primers, l μL of DMSO, 1 μL of 2.5 mM DNTPs, 0.1 μL of Taq DNA polymerase, 3 μL of 10 ng/μL genomic DNA and 13.4 μL nuclease free water. Amplification was done in a thermocycler with initial denaturation at 94°C for 5 min, followed by 36 cycles of denaturation at 94°C for 30 sec, annealing at 54°C for 30 sec and elongation at 72°C for 45 sec. Followed by a final elongation step at 72°C for 7 min and hold temperature at 10°C. Amplified fragments were visualized on safe view-stained 1.5% agarose gel.
The PCR products were sent to the International Institute of Tropical Agriculture (IITA), Ibadan for purification and sequencing. The PCR products were sequenced on ABI 3500 Genetic Analyzer (Thermo Fisher Scientific, Massachusetts, USA).
Phylogenetic analysis: Sequences were cleaned and identified on the BLAST algorithm on the National Centre for Biotechnology Information (NCBI) database. Best blast hits were used for the construction of a neighbor-joining phylogenetic tree which was used to determine the evolutionary relationship among the fungal species isolated and other species on GenBank. Evolutionary analysis was conducted on Molecular Evolutionary Genetics Analysis (MEGA) software, version 10.0.1 (MEGA X).
RESULTS
Fungal isolates of Telfairia occidentalis leaves: Two fungal organisms were isolated from Telfairia occidentalis leaves (Fig. 1a-b). The mean value of fungi colony on plated T. occidentalis leaves was calculated and the frequency of occurrence of each organism on PDA was determined. Isolate 1 had a higher frequency of occurrence (2.25) than isolate 2 which had 2.0.
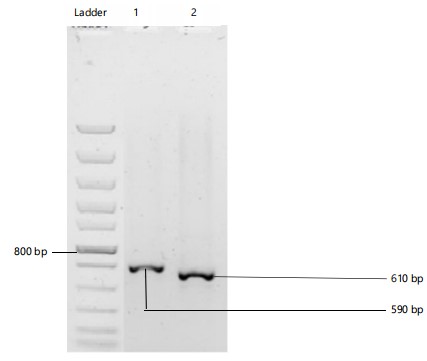
Polymerase Chain Reaction (PCR): The PCR profiles of isolates 1 and 2 showed bands on gel when viewed under UV light. The PCR product of each isolate is presented in Fig. 2.
ITS sequences obtained: Amplified PCR products after sequencing and blasting revealed the species identity of the fungal isolates to be Trichoderma reesei (isolate 1) and Aspergillus aculeatinus (isolate 2) in Fig. 2.
Accession numbers of isolates were obtained from GenBank and each fungal isolate was assigned a strain number. The accession numbers are in parentheses below:
| • | Isolate 1: Trichoderma reesei (ON965496) strain number RCBBR_AEAN9 | |
| • | Isolate 2: Aspergillus aculeatinus (ON965497) strain number RCBBR_AEAN10 |

|
|
|
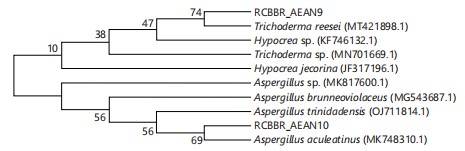
Phylogenetic analysis: The ITS sequences data of other fungal isolates evolutionarily related to the isolates obtained from T. occidentalis were obtained from GenBank and used to construct a phylogenetic tree. The closest relatives of the fungal isolates obtained from T. occidentalis are Hypocrea jecorina, Hypocrea sp., Trichoderma sp., Aspergillus brunneoviolaceus, A. trinidadensis and Aspergillus sp., as shown in Fig. 3.
DISCUSSION
The morphology of the isolated fungi was yellow to brown dark spores and a frequency of 2±0.71 and green sporulation and a frequency of 2.25±0.25 for Trichoderma reesei and Aspergillus aculeatinus, respectively. Molecular characterization tools offer insights into the identification and analysis of unknown species, allowing for the comparison of DNA sequences between known and unknown organisms. This molecular data enhances understanding of phylogeny and provides valuable information for studying the taxonomy and evolution of species. Molecular methods for identifying organisms are highly reliable, accurate, dependable and quick. Specifically, molecular characterization using Polymerase Chain Reaction (PCR) amplification and sequencing of internal transcribed spacer (ITS) regions was utilized to effectively identify fungal organisms isolated from Telferia occidentalis. This method identified two fungi Trichoderma reesei and Aspergillus aculeatinus.
Trichoderma reesei is a mesophilic and filamentous fungus that belongs to the phylum Ascomycota, class Sordariomycetes, order Hypocreales and family Hypocreales. Trichoderma reesei was originally isolated from Solomon Island during World War II because it degraded canvas and garments of the US army and all strains currently used in biotechnology and basic research were derived from this isolate17. Trichoderma reesei is an efficient producer of extracellular proteins hence it is widely used in cellulase and biofuel production. According to previous study of Bischof et al.18 Trichoderma reesei is a widely utilized organism in the biotechnology industry, serving as a key producer of cellulase enzymes. Its dominance as a workhorse in enzyme production has made it essential for various industrial applications. The fungus is also known to have antagonistic properties. Cellulases produced by T. reesei strongly inhibit Pepper Mild Mottle Virus infections in plants19. Trichoderma reesei was found to be an effective antagonistic microorganism against the plant pathogen, Rhizoctonia solani. Grosch et al.20 and Harish et al.21 Studied Bipolaris oryzae, which causes brown spot disease in rice. Trichoderma reesei is capable of mycoparasitism against Pythium ultimum, an aggressive soil-borne plant pathogen22. According to Sánchez-Montesinos et al.23, Trichoderma fungi can negatively impact certain areas of agriculture and human health. In agriculture, their adverse effects are primarily associated with their mycoparasitic capabilities, leading to a condition referred to as green mold, which poses a significant threat to the cultivation of mushrooms such as shiitake, oyster and champignons.
The genus Aspergillus is widely distributed across various ecosystems and substrates, including soil, textiles and food24. Molds like Aspergillus can negatively impact human health due to their potential to cause toxicity, trigger allergic reactions and lead to infections25. Aspergillus aculeatinus, a species within this genus, belongs to the group of black aspergilli and is closely related to Aspergillus aculeatus26. Through genome sequencing combined with phylogenetic and phenotypic analyses, A. aculeatinus was identified as a new species27. This species has been isolated from Thai coffee beans and shows potential for industrial use, producing bioactive compounds such as neoxaline, the antifungal compound aculeacin and the antitumor compound paclitaxel, initially known as Taxol by Bristol-Myers Squibb28. To date, only one genome of A. aculeatinus has been sequenced30. Species in Aspergillus section Nigri, which includes the black aspergilli, are known for causing food spoilage, plant diseases and for producing industrially significant compounds like lipases, amylase, citric acid and gluconic acid29. Due to the fermentation capabilities and high level of protein secretion in the solid-state culture of Aspergillus spp., it is often used by the industry for the production of pectin-degrading enzymes. Hence, A. aculeatinus has an extensive and highly conserved set of genes encoding cell wall degrading enzymes, although their precise biochemical characterization and specificities have to be determined28,30.
Morphological and microscopic characteristics form the foundation of traditional microbial identification techniques. However, this method can result in misidentification and is inadequate for pinpointing microorganisms at the species level. Molecular techniques, based on the genetic composition of organisms, provide precise identification since the genetic makeup of all living beings varies to some degree. Many scientists in developing countries primarily rely on traditional methods for identifying microorganisms, which can lead to incorrect interpretations of the fungal community. In this study, molecular techniques were successfully employed to identify fungal isolates from Telfairia occidentalis leaves. The study also highlights the ecology and economic significance of these fungal isolates, providing essential information for researchers to develop strategies for improving plant health.
CONCLUSION
Fungal species are associated with the T. occidentalis. Molecular characterization using Polymerase Chain Reaction (PCR) amplification and sequencing of internal transcribed spacer (ITS) regions proved effective in identifying fungal samples. This study will enhance understanding of the fungal species associated with Telfairia occidentalis and also aid plant pathologists make informed decisions on disease control resulting in a decrease in post-harvest loss and promoting crop protection.
SIGNIFICANCE STATEMENT
This study discovered the fungi species associated with postharvest Telfairia occidentalis leaves. Despite their significant nutritional benefits, Telfairia occidentalis leaves are compromised by pathogens that reduce their shelf life. Molecular identification of fungal organisms is crucial as it is faster, more reliable and more accurate, giving plant pathologists the knowledge needed to make informed decisions. Trichoderma reesei and Aspergillus. aculeatinus have been documented to cause spoilage and can be harmful to animals and humans when consumed. Proper storage and handling practices should be adopted to minimize fungal contamination and public awareness campaigns should be conducted to educate consumers on the potential health risks associated with consuming contaminated leaves.
ACKNOWLEDGMENT
The authors express special thanks to all the staff of the Regional Center for Biotechnology and Bio-resources Research, University of Port Harcourt, Rivers State, Nigeria for their assistance and contribution to the success of this research.
REFERENCES
- Ogbole, O.O., O.O. Abiodun and E.O. Ajaiyeoba, 2015. Antioxidant properties, macro and micro elements of selected edible vegetables. Niger. J. Pharm. Res., 11: 94-100.
- Akoroda, M.O., 1990. Ethnobotany of Telfairia occidentalis (cucurbitaceae) among Igbos of Nigeria. Econ. Bot., 44: 29-39.
- Uboh, F.E., M.I. Akpanabiatu, E.E. Edet and I.E. Okon, 2011. Distribution of heavy metals in fluted pumpkin (Telfeiria ocidentalis) leaves planted at different distances away from the traffic congested highways. Int. J. Adv. Biotechnol. Res., 2: 250-256.
- Ajayi, S.A., P. Berjak, J.I. Kioko, M.E. Dulloo and R.S. Vodouhe, 2006. Responses of fluted pumpkin (Telfairia occidentalis Hook. f.; Cucurbitaceae) seeds to desiccation, chilling and hydrated storage. S. Afr. J. Bot., 72: 544-550.
- Ubani, O.N. and E.U. Okonkwo, 2011. A review of shelf-life extension studies of Nigerian indigenous fresh fruits and vegetables in the Nigerian Stored Products Research Institute. Afr. J. Plant Sci., 5: 537-546.
- Temitope, O.R., O.O. Olugbenga, A.J. Erasmus, I. Jamilu and M.Y. Shehu, 2020. Comparative study of the physicochemical properties of male and female flutted pumpkin (Telfairia occidentalis). J. Med. Res., 6: 55-61.
- Osai, E.O., S.O. Akan and S.E. Udo, 2013. The efficacy of plantain inflorescence ash in the control of transluscent leafspot disease of Telfairia occidentalis (Hook F.). Int. J. Res. Appl. Nat. Social Sci., 1: 37-44.
- Obembe, O.M., D.O. Ojo and K.D. Ileke, 2021. Effect of different drying methods and storage on proximate and mineral composition of fluted pumpkin leaf, Telfairia occidentalis Hook f. J. Hortic. Postharvest Res., 4: 141-150.
- Nkiru, U., 2018. Antifungal effect and phytochemical screening of Telfairia occidentalis (hook f.) leaf extracts. J. Plant Biotechnol. Microbiol., 1: 21-24.
- Akwaowo, E.U., B.A. Ndon and E.U. Etuk, 2000. Minerals and antinutrients in fluted pumpkin (Telfairia occidentalis Hook f.). Food Chem., 70: 235-240.
- Egbekun, M.K. E.O. Nda-Suleiman and O. Akinyeye, 1998. Utilization of fluted pumpkin fruit (Telfairia occidentalis) in marmalade manufacturing. Plant Foods Hum. Nutr., 52: 171-176.
- Kimura, M. and D.B. Rodriguez-Amaya, 2003. Carotenoid composition of hydroponic leafy vegetables. J. Agric. Food Chem., 51: 2603-2607.
- Emeka, E.J.I. and O. Obidoa, 2009. Some biochemical, haematological and histological responses to a long term consumption of Telfairia occidentalis-supplemented diet in rats. Pak. J. Nutr., 8: 1199-1203.
- Mbong, G.A., A.K. Kebei, L.A. Agyingi, N.C.B. Tatiana, S.E. Mbong and N.E. Muluh, 2021. Influence of cropping system on the incidence and severity of leaf spot disease of Telfairia occidentalis Hook F. caused by Phoma sorghina. Int. J. Appl. Agric. Sci., 7: 162-168.
- Xue, K., M.M. Yuan, Z.J. Shi, Y. Qin and Y. Deng et al., 2016. Tundra soil carbon is vulnerable to rapid microbial decomposition under climate warming. Nat. Clim. Change, 6: 595-600.
- Long, P.E., K.H. Williams, S.S. Hubbard and J.F. Banfield, 2016. Microbial metagenomics reveals climate-relevant subsurface biogeochemical processes. Trends Microbiol., 24: 600-610.
- Seidl, V., C. Seibel, C.P. Kubicek and M. Schmoll, 2009. Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci., U.S.A., 106: 13909-13914.
- Bischof, R.H., J. Ramoni and B. Seiboth, 2016. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact., 15.
- Oka, N., T. Ohki, Y. Honda, K. Nagaoka and M. Takenaka, 2008. Inhibition of pepper mild mottle virus with commercial cellulases. J. Phytopathol., 156: 65-67.
- Grosch, R., K. Scherwinski, J. Lottmann and G. Berg, 2006. Fungal antagonists of the plant pathogen Rhizoctonia solani: Selection, control efficacy and influence on the indigenous microbial community. Mycol. Res., 110: 1464-1474.
- Harish, S., D. Saravanakumar, R. Radjacommare, E.G. Ebenezar and K. Seetharaman, 2008. Use of plant extracts and biocontrol agents for the management of brown spot disease in rice. BioControl, 53: 555-567.
- Monteiro, V.N., A.S. Steindorff, F.B. dos Reis Almeida, F.A.C. Lopes, C.J. Ulhoa, C.R. Félix and R.N. Silva, 2015. Trichoderma reesei mycoparasitism against Pythium ultimum is coordinated by G-alpha protein GNA1 signaling. J. Microb. Biochem. Technol., 7: 1-7.
- Sánchez-Montesinos, B., M. Santos, A. Moreno-Gavíra, T. Marín-Rodulfo, F.J. Gea and F. Diánez, 2021. Biological control of fungal diseases by Trichoderma aggressivum f. europaeum and its compatibility with fungicides. J. Fungi, 7.
- Klich, M.A., 2009. Health effects of Aspergillus in food and air. Toxicol. Ind. Health, 25: 657-667.
- Hedayati, M.T., S. Mayahi and D.W. Denning, 2010. A study on Aspergillus species in houses of asthmatic patients from Sari City, Iran and a brief review of the health effects of exposure to indoor Aspergillus. Environ. Monit. Assess., 168: 481-487.
- Noonim, P., W. Mahakarnchanakul, J. Varga, J.C. Frisvad and R.A. Samson, 2008. Two novel species of Aspergillus section Nigri from Thai coffee beans. Int. J. Syst. E Microbiol., 58: 1727-1734.
- Qiao, W., T. Tang and F. Ling, 2020. Comparative transcriptome analysis of a taxol-producing endophytic fungus, Aspergillus aculeatinus Tax-6, and its mutant strain. Sci. Rep., 10.
- Vesth, T.C., J.L. Nybo, S. Theobald, J.C. Frisvad and T.O. Larsen et al., 2018. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat. Genet., 50: 1688-1695.
- Varga, J., J.C. Frisvad, S. Kocsubé, B. Brankovics, B. Tóth, G. Szigeti and R.A. Samson, 2011. New and revisited species in Aspergillus section Nigri. Stud. Mycol., 69: 1-17.
- Lemaire, A., C.D. Garzon, A. Perrin, O. Habrylo and P. Trezel et al., 2020. Three novel rhamnogalacturonan I- pectins degrading enzymes from Aspergillus aculeatinus: Biochemical characterization and application potential. Carbohydr. Polym., 248.
How to Cite this paper?
APA-7 Style
Gloria,
O.N., Obaraemi,
T.J., Basene,
J.O. (2024). Identification of Post Harvest Fungi Associated Telfairia occidentalis Hook F. Leaves Using Basic Molecular Techniques
. Research Journal of Botany, 19(1), 84-91. https://doi.org/10.3923/rjb.2024.84.91
ACS Style
Gloria,
O.N.; Obaraemi,
T.J.; Basene,
J.O. Identification of Post Harvest Fungi Associated Telfairia occidentalis Hook F. Leaves Using Basic Molecular Techniques
. Res. J. Bot 2024, 19, 84-91. https://doi.org/10.3923/rjb.2024.84.91
AMA Style
Gloria
ON, Obaraemi
TJ, Basene
JO. Identification of Post Harvest Fungi Associated Telfairia occidentalis Hook F. Leaves Using Basic Molecular Techniques
. Research Journal of Botany. 2024; 19(1): 84-91. https://doi.org/10.3923/rjb.2024.84.91
Chicago/Turabian Style
Gloria, Ogbuji, Nkechi, Tariah Julia Obaraemi, and Jonah Obedo Basene.
2024. "Identification of Post Harvest Fungi Associated Telfairia occidentalis Hook F. Leaves Using Basic Molecular Techniques
" Research Journal of Botany 19, no. 1: 84-91. https://doi.org/10.3923/rjb.2024.84.91

This work is licensed under a Creative Commons Attribution 4.0 International License.



