Studies on Genetic Polymorphism of Improved Cowpea Varieties Using Simple Sequence Repeats (SSR) Marker
| Received 19 Apr, 2023 |
Accepted 03 Jul, 2023 |
Published 06 Jul, 2023 |
Background and Objective: Several new varieties are released from breeding programs targeted at solving specific threats to cowpea production in Nigeria. As part of efforts to promote crop improvement, this study was carried out to determine the level of genetic polymorphism and phylogenetic relationship that exists among four selected improved varieties of cowpea (SAMPEA-14, SAMPEA-15, SAMPEA-17 and SAMPEA-18) using ten SSR molecular markers. Materials and Methods: The DNA was extracted from 14 days old seedling using the CTAB method. A total of 10 SSR primers were used in the DNA amplification process on a programmed thermal cycler followed by electrophoresis, visualization and scoring of the banding pattern. Data were analyzed on the Minitab 16.0 software for clustering patterns while the Polymorphic Information Content (PIC) of each primer was calculated. Results: Polymorphic information content (PIC) ranged from 0.619 in RB20 to 0.881 in RB38 primers. The top three markers in PIC values were RB38 (PIC = 0.881), CLM0342 (PIC = 0.873) and RB7 (PIC = 0.866). The mean PIC of the ten primers was 0.884. The genetic similarity index of the dendrogram was very low as SAMPEA-17 showed the lowest similarity coefficients of 18.35 away from other varieties while SAMPEA-15 had similarity coefficients of 33.33. Conclusion: This study has revealed high genetic differences among the four varieties studied. All of the varieties may be used as genetic materials in breeding work to improve local landraces to achieve sustainable cowpea production and food security in Nigeria.
| Copyright © 2023 Joseph et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Cowpea (Vigna unguiculata (L.) Walp) has become an important food crop being an essential diet in many developing countries including Africa and Asia1. With its origin first traced to Africa, its cultivation spreads throughout the tropical and semi-arid regions of the world2. It serves as a means of livelihood that generates income for the farmers and those who trade on the crop. It is the cheapest source of protein when compared to fish, meat and egg3. After the harvest of the grains, the fodder serves as food for livestock4.
Cowpea is predominantly grown in Africa with Nigeria and Niger Republic as leading producers. Brazil, West Indies, India, Srilanka, Yugoslavia and Australia are also producing countries4. The different habits of growth identified are erect, prostrate, climbing and glabrous3. The crop is suitable for drought-prone and heat-generating regions like ours (Nigeria) where other food legumes fail due to inadequate rains. It can fix atmospheric nitrogen through its root noodles and as a result, thrives in nutrient-deficient soils where other crop fails. This feature, in addition to its ability to survive shady areas, makes cowpea an intercrop-friendly crop with cereals like maize, millet and sorghum5 where the fixed nitrogen is also utilized by crops that are intercropped with or grown in rotation with cowpea. Despite the valuable nature of cowpea, its production is constrained by a lot of biotic and abiotic factors1. Efforts to improve the crop are of utmost importance and many research institutions, either singly or through collaborative means, have deployed different approaches such as the conventional breeding methods or use of molecular markers in marker-assisted selection (MAS) and marker-assisted breeding (MAB) respectively to solve the problem limiting production1. Molecular markers are a valuable and reliable tool that have accelerated different research programs and are effective in cost and labor while maintaining outstanding accuracy6. Simple sequence repeat (SSR) markers have been applied in genetic diversity studies of cowpea7-10. It has been used in the determination of phylogenetic relationships among cowpea genotypes11 and in the population genetic studies of other crops12.
The different cowpea varieties at the molecular Biology Laboratory of Joseph Sarwuan Tarka University, Makurdi are the most accessible part of cowpea diversity by farmers around the university community, Benue State and some parts of Nigeria. Several new varieties are released from breeding programs that were targeted at solving the problems faced by cowpea farmers in the community, state and Nigeria at large. However, the genetic knowledge about these varieties which could be used to understand their evolutionary relationship is still lacking. This knowledge gap is a serious limitation to utilizing, managing and conserving the cowpea gene pool in the university. The use of morphological markers to explain evolutionary relationships would have been easy but may be misleading due to their epistatic and pleiotropic effects. Morphological markers are also affected by environmental conditions and the stage of growth of the plants, this makes them unreliable. Molecular markers, however, are efficient in genetic and phylogenetic studies as reported by Mirzaei6. Olasupo et al.9 encouraged phylogenetic studies on new varieties using genetic-based tools and molecular techniques. This study aimed at determining the level of genetic polymorphism and phylogenetic relationship that exists among four selected improved varieties of cowpea (SAMPEA-14, SAMPEA-15, SAMPEA-17 and SAMPEA-18) using SSR molecular markers.
MATERIALS AND METHODS
Collection of samples: The study was carried out from August, 2022 to November, 2022. Twenty seeds each of the four improved varieties of cowpea (SAMPEA-14, SAMPEA-15, SAMPEA-17 and SAMPEA-18) were obtained from the Molecular Biology Laboratory of the Department of Plant Breeding and Seed Science, University of Agriculture Makurdi, Benue State, Nigeria.
Planting for DNA extraction: Seeds were planted in rubber pots in the Laboratory Screen House where breeding work is usually done. They were watered at intervals. After two weeks of planting, leaves of seedlings were collected in leather sachets containing silica gel and preserved in a desiccator to enable them dry. After 4 days, the leaves were dried and were due for extraction of DNA.
DNA extraction: The CTAB method of DNA extraction was used13,14. Two leaves of a trifoliate 14 days old plant were placed in silica gel for 3 days to dry. The crispy dry leaves were squeezed into a 2 mL Eppendorf tube containing two steel balls (leave tissue is about 1/6 of 2 mL Eppendorf tube) and ground vigorously using a vortex for 1 min until it becomes powder. Buffer (1M Tris-HCL, 0.5M EDTA, 5M NaCl, 2-Mecarptoethanol) was added and incubated in a water bath for 30 min at 60°C. As 600 μL of Chloroform: Isoamylalcohol (24:1) was added and spun for 10 min at 4000 rpm. The upper layer was transferred into new tubes and this step was repeated. As 600 μL of ice-cold 2-propanol was added into the supernatant and inverted for a few seconds. Tubes were kept at -20°C overnight to precipitate nucleic acid out of the solution. Tubes were then centrifuged for 35 min at 4000 rpm to form a pellet at the bottom of the tube and the supernatant was discarded. The pellet was washed with 400 μL of 70% ethanol, centrifuged for 15 min and ethanol decanted. The process was repeated and the pellet air dried for about 1 hr (until no droplet of ethanol was seen). The pellet was suspended in 100 mL of molecular-grade water/RNase water. The quality was checked using 0.8% Agarose gel13.
| Table 1: | Polymorphic information content of SSR Primers | |||
| Primer name | Sequence | Pi |
PIC 1-Σpi2 |
| RB38 | GCGGCCGCTGCTCGTTCCCG | 0.345 |
0.881 |
| CLM0775 | GTGGCAGCACAAGTTAGTAG | 0.418 |
0.825 |
| RB43 | CCATGGTCGCCCCTGCTGCACCTTG | 0.518 |
0.732 |
| RB18 | AAGCTTACTTGTACAGCTCGTCCATGCCG | 0.367 |
0.865 |
| CLM1190 | ATTTGGCTGAATTGTTTCCA | 0.487 |
0.763 |
| CLM0400 | CATGGTGTACAGATTTGTGG | 0.366 |
0.866 |
| RB7 | GGGCGTTAATTAAGCCCACACA | 0.519 |
0.731 |
| RB20 | CCATGGGGGCATCAACCTTGG | 0.617 |
0.619 |
| CLM0342 | GGATTGGATATGTGTCTGGC | 0.357 |
0.873 |
| CLM0218 | TTTCCGATTTGCGATTTTTA | 0.445 |
0.802 |
| Total | 4.439 |
7.957 |
|
| Mean | 0.493 |
0.884 |
PCR analysis: The PCR (polymerase chain reaction) was carried out in a thermal cycler (Applied Bio system in Life Technology 2720 Model) in 35 cycles consisting of the following gradient profiles: Denaturation at 94°C for the 30 sec, annealing at 57.5°C for 30 sec, extension at 72°C for 2 min and a final hold at 72°C for 10 min using the method of Omoigui et al.13,15. Each SSR primer was applied for screening all the varieties for PCR amplification. The list of 10 SSR primers used in the amplification and their sequence was given in Table 1.
Gel electrophoresis: Amplicons were dispensed into the agarose gel-based electrophoresis chamber (Galileo Bioscience tank connected to Consort EV243 electrophoresis power supply) that consisted of 1X Tris acetic acid (TAE) buffer at 8.4 pH. A 50 kb ladder as the reference band ethidium bromide was also dispensed to stain the DNA for visualization. The connection ran at 120 Volts for 90 min. The separated bands were visualized on a UV trans illuminator while photographs were taken using a camera15 (Canon SX120).
Scoring of gel images and statistical analysis: The DNA bands were scored according to the method of Aguoru et al.16 to generate a binary matrix. This was uploaded on the Minitab 17.0 software. Polymorphic bands were analyzed for each SSR primer used. Calculation of polymorphic information content (PIC) was done using the formula14:
PIC = 1- Σpi2 |
Cluster analysis was done using the Single Linkage method measured on Euclidean Distance. The level of significance was set at 5% (p≤0.05).
RESULTS AND DISCUSSION
Selected gel images of the DNA of the four cowpea varieties amplified by SSR primers were shown in Plate 1. Primers gave a clear resolution of bands that indicate the presence of marker genes coding for a particular trait, possibly disease resistance genes. This was because the selected primers were previously used in cowpea breeding for resistance to diseases17.

|

|
The polymorphic information content (PIC) of the SSR primers was given in Table 1. It ranged from 0.619 in RB20 to 0.881 in RB38 primers. The top four in PIC values were RB38 (PIC = 0.881), CLM0342 (PIC = 0.873) CLM0400 (PIC = 0.866) and RB18 (0.865). The mean PIC of the ten primers was 0.884. The reported PICs were higher than the average of 0.51 earlier given by Olasupo et al.9 in the SSR markers for evaluation of genetic diversity in mutant cowpea lines. In their studies, primers used were considered polymorphic since they surpassed the 0.50 benchmark. Ogunkanmi et al.8 in their studies on cowpea lines characterized by 12 SSR markers described the genotypes as high in genetic difference as PIC was between 0.603 and 0.705. Thus, the four varieties of cowpea studies in this work are very high in terms of genetic differences as indicated by a maximum PIC of 0.881.
SAMPEA-15 and SAMPEA-17 had the highest percentage number of bands of 30% each amplified by all primers. SAMPEA-18 recorded 25% while SAMPEA-14 was the least (15%) as shown in Fig. 1. These bands are DNA macromolecules carrying the genes flanked by the marker under study, hence, unique traits of interest are identified. This submission is in tandem with other reports where SSR markers are used as landmarks for monitoring traits13 and constructing the genetic structure of a population of Vigna unguiculata12.
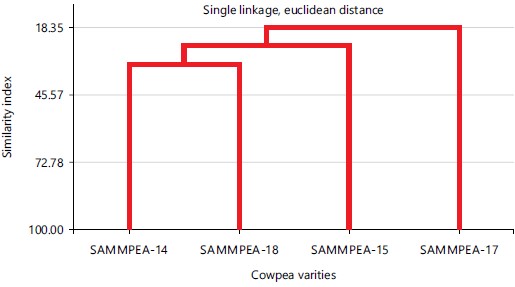
The information provided by the ten SSR markers is complimentary to the results of cluster analysis that gave very high genetic differences. The genetic similarity index of the dendrogram was very low as SAMPEA-17 showed the lowest similarity coefficients of 18.35 away from other varieties while SAMPEA-15 had similarity coefficients of 33.33 as shown in Fig. 2. These are the most distant genotypes that diverged from the other two varieties. SAMPEA-14 and SAMPEA-18 are closely related in genetic constituents based on the primers employed. A similar pattern of genetic divergence was earlier presented by Badiane et al.11 using 49 SSR markers to determine the phylogenetic relationship that exists among some local varieties and inbred lines. It also supported recent phylogenetic constructions among some legumes using SSR markers6,18.
The outcome of this study implies that there are genetic differences among the four varieties of cowpea where sample-17 was an outstanding genotype based on the molecular marker employed. This could be a reflection of differences in genetic coding for a useful agronomic trait. These genetic materials could be applied in breeding work to improve other varieties. A high difference between two sequences usually implies significant functional or structural divergence and these sequences are unrelated in a phylogenetic tree. This study recommends further molecular studies on the four varieties of cowpea using extensive molecular breeding tools as the present work is limited by the size of the markers employed. Hence, a large number of SSR and other markers may be used in future work.
|
CONCLUSION
This study has revealed high genetic differences among the four varieties studied. Polymorphic information content values of the 10 SSR primers were very high thus complimenting the low genetic similarity coefficients reported. SAMPEA-17 was the most distant of the four varieties. All of the varieties may be used as genetic materials in breeding work to improve local landraces to achieve sustainable cowpea production and food security in Nigeria.
SIGNIFICANCE STATEMENT
Several new varieties are released from breeding programs targeted at solving specific threats to sufficient cowpea production in Nigeria. As part of efforts to promote crop improvement, this study was carried out to determine the level of genetic polymorphism and phylogenetic relationship that exists among four selected improved varieties of cowpea (SAMPEA-14, SAMPEA-15, SAMPEA-17 and SAMPEA-18) using 10 SSR molecular markers.
REFERENCES
- Olasan, O.J., C.U. Aguoru, L. Omoigui, F. Oluma and M.S. Ugbaa et al., 2023. Genetic diversity and phylogenetics of four released cowpea (Vigna unguiculata (L.) Walp) varieties (fuampea-1, fuampea-2, fuampea-3 and fuampea-4) using simple sequence repeats markers. J. Exp. Mol. Biol., 24: 41-50.
- Puozaa, D.K., S.K. Jaiswal and F.D. Dakora, 2019. Phylogeny and distribution of Bradyrhizobium symbionts nodulating cowpea (Vigna unguiculata L. Walp) and their association with the physicochemical properties of acidic African soils. Syst. Appl. Microbiol., 42: 403-414.
- Xiong, H., A. Shi, B. Mou, J. Qin and D. Motes et al., 2016. Genetic diversity and population structure of cowpea (Vigna unguiculata L. Walp). PLoS ONE, 11: 0160941.
- Badr, A., H.I.S. Ahmed, M. Hamouda, M. Halawa and M. Elhiti, 2014. Variation in growth, yield and molecular genetic diversity of M2 plants of cowpea following exposure to gamma radiation. Life Sci. J., 11: 10-19.
- Iqbal, M.A., 2015. Improving germination and seedling vigour of cowpea (Vigna unguiculata L.) with different priming techniques. Am. Eurasian J. Agric. Environ. Sci., 15: 265-270.
- Mirzaei, S., 2021. Application of molecular markers in plant sciences; An overview. Cent. Asian J. Plant Sci. Innovation, 4: 192-200.
- Asare, A.T., B.S. Gowda, I.K.A. Galyuon, L.L. Aboagye, J.F. Takrama and M.P. Timko, 2010. Assessment of the genetic diversity in cowpea (Vigna unguiculata L. Walp.) germplasm from Ghana using simple sequence repeat markers. Plant Genet. Resour., 8: 142-150.
- Ogunkanmi, L.A., O.T. Ogundipe and C.A. Fatokun, 2014. Molecular characterization of cultivated cowpea (Vigna unguiculata L. Walp) using simple sequence repeats markers. Afr. J. Biotechnol., 13: 3464-3472.
- Olasupo, F.O., C.O. Ilori, B.P. Forster and S. Bado, 2018. Selection for novel mutations induced by gamma irradiation in cowpea [Vigna unguiculata (L.) Walp.]. Int. J. Plant Breed. Genet., 12: 1-12.
- Aguoru, C.U., J.O. Olasan, L.O. Omoigui and E.J. Ekefan, 2022. Marker assisted identification of suitable candidates for biosystematics and crop improvement among groundnut (Arachis hypogaea L.) breeding lines. Nigeria J. Plant Breed., 1: 43-54.
- Badiane, F.A., B.S. Gowda, N. Cissé, D. Diouf, O. Sadio and M.P. Timko, 2012. Genetic relationship of cowpea (Vigna unguiculata) varieties from Senegal based on SSR markers. Genet. Mol. Res., 11: 292-304.
- Kanyika, B.T.N., D. Lungu, A.M. Mweetwa, E. Kaimoyo and V.M. Njung'e et al., 2015. Identification of groundnut (Arachis hypogaea) SSR markers suitable for multiple resistance traits QTL mapping in African germplasm. Electr. J. Biotechnol., 18: 61-67.
- Omoigui, L.O, M.F. Ishiyaku, B.S. Gowda, A.Y. Kamara and M.P. Timko, 2015. Suitability and use of two molecular markers to track race-specific resistance Striga gesnerioides in cowpea (Vigna unguiculata (L.) Walp.). Afr. J. Biotechnol., 14: 2179-2190.
- Tersoo, K., I.C. Chidozie, O.L. Osabuohien, O.J. Olalekan and U.M. Sesugh, 2021. Characterization of some selected tomato (Solanum lycorpersicum L) accessions using simple sequence repeat markers. Agric. Biol. Sci. J., 7: 46-50.
- Omoigui, L.O., A.Y. Kamara, G.I. Alunyo, L.L. Bello, M. Oluoch, M.P. Timko and O. Boukar, 2017. Identification of new sources of resistance to Striga gesnerioides in cowpea Vigna unguiculata accessions. Genet. Resour. Crop Evol., 64: 901-911.
- Aguoru, C.U., L.O. Omoigui and J.O. Olasan, 2015. Population genetic study of eggplants (Solanum) species in Nigeria, Tropical West Africa, using molecular markers. Int. J. Plant Res., 5: 7-12.
- Omoigui, L.O., A.Y. Kamara, Y.D. Moukoumbi, L.A. Ogunkanmi and M.P. Timko, 2017. Breeding cowpea for resistance to Striga gesnerioides in the Nigerian dry savannas using marker-assisted selection. Plant Breed., 136: 393-399.
- Omoigui, L.O., G.C. Ekeuro, A.Y. Kamara, L.L. Bello, M.P. Timko and G.O. Ogunwolu, 2017. New sources of aphids [Aphis craccivora (Koch)] resistance in cowpea germplasm using phenotypic and molecular marker approaches. Euphytica, 213: 178.
How to Cite this paper?
APA-7 Style
Joseph,
O.O., Osabuohien,
O.L., Apeh,
O.H., Uzoma,
A.C., Dennis,
D., Sesugh,
U.M., Judith,
E., Godspower,
E., Paul,
D., Nater,
I., Mnzughul,
S., Thomas,
O. (2023). Studies on Genetic Polymorphism of Improved Cowpea Varieties Using Simple Sequence Repeats (SSR) Marker. Research Journal of Botany, 18(1), 36-42. https://doi.org/10.3923/rjb.2023.36.42
ACS Style
Joseph,
O.O.; Osabuohien,
O.L.; Apeh,
O.H.; Uzoma,
A.C.; Dennis,
D.; Sesugh,
U.M.; Judith,
E.; Godspower,
E.; Paul,
D.; Nater,
I.; Mnzughul,
S.; Thomas,
O. Studies on Genetic Polymorphism of Improved Cowpea Varieties Using Simple Sequence Repeats (SSR) Marker. Res. J. Bot 2023, 18, 36-42. https://doi.org/10.3923/rjb.2023.36.42
AMA Style
Joseph
OO, Osabuohien
OL, Apeh
OH, Uzoma
AC, Dennis
D, Sesugh
UM, Judith
E, Godspower
E, Paul
D, Nater
I, Mnzughul
S, Thomas
O. Studies on Genetic Polymorphism of Improved Cowpea Varieties Using Simple Sequence Repeats (SSR) Marker. Research Journal of Botany. 2023; 18(1): 36-42. https://doi.org/10.3923/rjb.2023.36.42
Chicago/Turabian Style
Joseph, Olasan, Olalekan, Omoigui Lucky Osabuohien, Oluma Hyacinth Apeh, Aguoru Celestine Uzoma, Deo Dennis, Ugbaa Macsamuel Sesugh, Ezugwu Judith, Ekeruo Godspower, Dughduh Paul, Iyorkaa Nater, Simon Mnzughul, and Okoh Thomas.
2023. "Studies on Genetic Polymorphism of Improved Cowpea Varieties Using Simple Sequence Repeats (SSR) Marker" Research Journal of Botany 18, no. 1: 36-42. https://doi.org/10.3923/rjb.2023.36.42

This work is licensed under a Creative Commons Attribution 4.0 International License.



